Organismo modello: differenze tra le versioni
→Vertebrati: testo |
→Vertebrati: ins. ref. |
||
| Riga 115: | Riga 115: | ||
:* [[Zebrafish|''Brachydanio rerio'']]<ref name="Kocher-2005">{{Cite journal | last1 = Kocher | first1 = TD. | last2 = Jeffery | first2 = WR. | last3 = Parichy | first3 = DM. | last4 = Peichel | first4 = CL. | last5 = Streelman | first5 = JT. | last6 = Thorgaard | first6 = GH. | title = Special feature--roundtable discussion. Fish models for studying adaptive evolution and speciation. | journal = Zebrafish | volume = 2 | issue = 3 | pages = 147-56 | month = | year = 2005 | doi = 10.1089/zeb.2005.2.147 | PMID = 18248189 }}</ref><ref name="Bradbury-2004">{{Cite journal | last1 = Bradbury | first1 = J. | title = Small fish, big science. | journal = PLoS Biol | volume = 2 | issue = 5 | pages = E148 | month = May | year = 2004 | doi = 10.1371/journal.pbio.0020148 | PMID = 15138510 }}</ref> o (''Danio rerio'') conosciuto come ''[[Zebrafish]]'', è un pesce d'acqua dolce molto usato negli acquari. Ha un corpo quasi trasparente durante il primo sviluppo, ciò fornisce un accesso visivo all'anatomia interna dell'animale. Gli Zebrafish sono utilizzati per studiare lo sviluppo, la tossicologia<ref name="Hill-2005">{{Cite journal | last1 = Hill | first1 = AJ. | last2 = Teraoka | first2 = H. | last3 = Heideman | first3 = W. | last4 = Peterson | first4 = RE. | title = Zebrafish as a model vertebrate for investigating chemical toxicity. | journal = Toxicol Sci | volume = 86 | issue = 1 | pages = 6-19 | month = Jul | year = 2005 | doi = 10.1093/toxsci/kfi110 | PMID = 15703261 }}</ref> e tossicopatologia<ref name="Spitsbergen-">{{Cite journal | last1 = Spitsbergen | first1 = JM. | last2 = Kent | first2 = ML. | title = The state of the art of the zebrafish model for toxicology and toxicologic pathology research--advantages and current limitations. | journal = Toxicol Pathol | volume = 31 Suppl | issue = | pages = 62-87 | month = | year = | doi = | PMID = 12597434 }}</ref> e la specifica funzione del gene e il ruolo delle vie di segnalazione. |
:* [[Zebrafish|''Brachydanio rerio'']]<ref name="Kocher-2005">{{Cite journal | last1 = Kocher | first1 = TD. | last2 = Jeffery | first2 = WR. | last3 = Parichy | first3 = DM. | last4 = Peichel | first4 = CL. | last5 = Streelman | first5 = JT. | last6 = Thorgaard | first6 = GH. | title = Special feature--roundtable discussion. Fish models for studying adaptive evolution and speciation. | journal = Zebrafish | volume = 2 | issue = 3 | pages = 147-56 | month = | year = 2005 | doi = 10.1089/zeb.2005.2.147 | PMID = 18248189 }}</ref><ref name="Bradbury-2004">{{Cite journal | last1 = Bradbury | first1 = J. | title = Small fish, big science. | journal = PLoS Biol | volume = 2 | issue = 5 | pages = E148 | month = May | year = 2004 | doi = 10.1371/journal.pbio.0020148 | PMID = 15138510 }}</ref> o (''Danio rerio'') conosciuto come ''[[Zebrafish]]'', è un pesce d'acqua dolce molto usato negli acquari. Ha un corpo quasi trasparente durante il primo sviluppo, ciò fornisce un accesso visivo all'anatomia interna dell'animale. Gli Zebrafish sono utilizzati per studiare lo sviluppo, la tossicologia<ref name="Hill-2005">{{Cite journal | last1 = Hill | first1 = AJ. | last2 = Teraoka | first2 = H. | last3 = Heideman | first3 = W. | last4 = Peterson | first4 = RE. | title = Zebrafish as a model vertebrate for investigating chemical toxicity. | journal = Toxicol Sci | volume = 86 | issue = 1 | pages = 6-19 | month = Jul | year = 2005 | doi = 10.1093/toxsci/kfi110 | PMID = 15703261 }}</ref> e tossicopatologia<ref name="Spitsbergen-">{{Cite journal | last1 = Spitsbergen | first1 = JM. | last2 = Kent | first2 = ML. | title = The state of the art of the zebrafish model for toxicology and toxicologic pathology research--advantages and current limitations. | journal = Toxicol Pathol | volume = 31 Suppl | issue = | pages = 62-87 | month = | year = | doi = | PMID = 12597434 }}</ref> e la specifica funzione del gene e il ruolo delle vie di segnalazione. |
||
:* [[Taeniopygia guttata]]<ref name="Benskin-2010">{{Cite journal | last1 = Benskin | first1 = CM. | last2 = Rhodes | first2 = G. | last3 = Pickup | first3 = RW. | last4 = Wilson | first4 = K. | last5 = Hartley | first5 = IR. | title = Diversity and temporal stability of bacterial communities in a model passerine bird, the zebra finch. | journal = Mol Ecol | volume = | issue = | pages = | month = Nov | year = 2010 | doi = 10.1111/j.1365-294X.2010.04892.x | PMID = 21054607 }}</ref><ref name="Warren-2010">{{Cite journal | last1 = Warren | first1 = WC. | last2 = Clayton | first2 = DF. | last3 = Ellegren | first3 = H. | last4 = Arnold | first4 = AP. | last5 = Hillier | first5 = LW. | last6 = Künstner | first6 = A. | last7 = Searle | first7 = S. | last8 = White | first8 = S. | last9 = Vilella | first9 = AJ. | title = The genome of a songbird. | journal = Nature | volume = 464 | issue = 7289 | pages = 757-62 | month = Apr | year = 2010 | doi = 10.1038/nature08819 | PMID = 20360741 }}</ref><ref name="Luo-2006">{{Cite journal | last1 = Luo | first1 = M. | last2 = Yu | first2 = Y. | last3 = Kim | first3 = H. | last4 = Kudrna | first4 = D. | last5 = Itoh | first5 = Y. | last6 = Agate | first6 = RJ. | last7 = Melamed | first7 = E. | last8 = Goicoechea | first8 = JL. | last9 = Talag | first9 = J. | title = Utilization of a zebra finch BAC library to determine the structure of an avian androgen receptor genomic region. | journal = Genomics | volume = 87 | issue = 1 | pages = 181-90 | month = Jan | year = 2006 | doi = 10.1016/j.ygeno.2005.09.005 | PMID = 16321505 }}</ref> (Zebra finch) o (diamante mandarino viene usato per studi sugli apparati uditivi dei non mammiferi. |
:* [[Taeniopygia guttata]]<ref name="Benskin-2010">{{Cite journal | last1 = Benskin | first1 = CM. | last2 = Rhodes | first2 = G. | last3 = Pickup | first3 = RW. | last4 = Wilson | first4 = K. | last5 = Hartley | first5 = IR. | title = Diversity and temporal stability of bacterial communities in a model passerine bird, the zebra finch. | journal = Mol Ecol | volume = | issue = | pages = | month = Nov | year = 2010 | doi = 10.1111/j.1365-294X.2010.04892.x | PMID = 21054607 }}</ref><ref name="Warren-2010">{{Cite journal | last1 = Warren | first1 = WC. | last2 = Clayton | first2 = DF. | last3 = Ellegren | first3 = H. | last4 = Arnold | first4 = AP. | last5 = Hillier | first5 = LW. | last6 = Künstner | first6 = A. | last7 = Searle | first7 = S. | last8 = White | first8 = S. | last9 = Vilella | first9 = AJ. | title = The genome of a songbird. | journal = Nature | volume = 464 | issue = 7289 | pages = 757-62 | month = Apr | year = 2010 | doi = 10.1038/nature08819 | PMID = 20360741 }}</ref><ref name="Luo-2006">{{Cite journal | last1 = Luo | first1 = M. | last2 = Yu | first2 = Y. | last3 = Kim | first3 = H. | last4 = Kudrna | first4 = D. | last5 = Itoh | first5 = Y. | last6 = Agate | first6 = RJ. | last7 = Melamed | first7 = E. | last8 = Goicoechea | first8 = JL. | last9 = Talag | first9 = J. | title = Utilization of a zebra finch BAC library to determine the structure of an avian androgen receptor genomic region. | journal = Genomics | volume = 87 | issue = 1 | pages = 181-90 | month = Jan | year = 2006 | doi = 10.1016/j.ygeno.2005.09.005 | PMID = 16321505 }}</ref> (Zebra finch) o (diamante mandarino viene usato per studi sugli apparati uditivi dei non mammiferi. |
||
:* [[Macaca mulatta]]<ref name="Snowden-1998">{{Cite journal | last1 = Snowden | first1 = KF. | last2 = Didier | first2 = ES. | last3 = Orenstein | first3 = JM. | last4 = Shadduck | first4 = JA. | title = Animal models of human microsporidial infections. | journal = Lab Anim Sci | volume = 48 | issue = 6 | pages = 589-92 | month = Dec | year = 1998 | doi = | PMID = 10090081 }}</ref> - Il Rhesus macaque o macaco vien usato negli studi sulla cognizione<ref name="Sripati-2010">{{Cite journal | last1 = Sripati | first1 = AP. | last2 = Olson | first2 = CR. | title = Global image dissimilarity in macaque inferotemporal cortex predicts human visual search efficiency. | journal = J Neurosci | volume = 30 | issue = 4 | pages = 1258-69 | month = Jan | year = 2010 | doi = 10.1523/JNEUROSCI.1908-09.2010 | PMID = 20107054 }}</ref><ref name="Samonds-2009">{{Cite journal | last1 = Samonds | first1 = JM. | last2 = Potetz | first2 = BR. | last3 = Lee | first3 = TS. | title = Cooperative and competitive interactions facilitate stereo computations in macaque primary visual cortex. | journal = J Neurosci | volume = 29 | issue = 50 | pages = 15780-95 | month = Dec | year = 2009 | doi = 10.1523/JNEUROSCI.2305-09.2009 | PMID = 20016094 }}</ref><ref name="Nekovarova-2009">{{Cite journal | last1 = Nekovarova | first1 = T. | last2 = Nedvidek | first2 = J. | last3 = Klement | first3 = D. | last4 = Bures | first4 = J. | title = Spatial decisions and cognitive strategies of monkeys and humans based on abstract spatial stimuli in rotation test. | journal = Proc Natl Acad Sci U S A | volume = 106 | issue = 36 | pages = 15478-82 | month = Sep | year = 2009 | doi = 10.1073/pnas.0907053106 | PMID = 19706408 }}</ref><ref name="Adachi-2009">{{Cite journal | last1 = Adachi | first1 = I. | last2 = Chou | first2 = DP. | last3 = Hampton | first3 = RR. | title = Thatcher effect in monkeys demonstrates conservation of face perception across primates. | journal = Curr Biol | volume = 19 | issue = 15 | pages = 1270-3 | month = Aug | year = 2009 | doi = 10.1016/j.cub.2009.05.067 | PMID = 19559613 }}</ref> e nelle malattie infettive.<ref name="Wilson-2009">{{Cite journal | last1 = Wilson | first1 = NA. | last2 = Watkins | first2 = DI. | title = Is an HIV vaccine possible? | journal = Braz J Infect Dis | volume = 13 | issue = 4 | pages = 304-10 | month = Aug | year = 2009 | doi = | PMID = 20231996 }}</ref><ref name="Silvestri-2009">{{Cite journal | last1 = Silvestri | first1 = G. | title = Immunity in natural SIV infections. | journal = J Intern Med | volume = 265 | issue = 1 | pages = 97-109 | month = Jan | year = 2009 | doi = 10.1111/j.1365-2796.2008.02049.x | PMID = 19093963 }}</ref><ref name="Grimaldi Jr-2008">{{Cite journal | last1 = Grimaldi Jr | first1 = G. | title = The utility of rhesus monkey (Macaca mulatta) and other non-human primate models for preclinical testing of Leishmania candidate vaccines. | journal = Mem Inst Oswaldo Cruz | volume = 103 | issue = 7 | pages = 629-44 | month = Nov | year = 2008 | doi = | PMID = 19057811 }}</ref> |
|||
:* [[Macaca mulatta]] - Il Rhesus macaque o macaco vien usato negli studi sulla cognizione e nelle malattie infettive. |
|||
:* [[Felis sylvestris catus]] - Il gatto è usato nelle ricerche di neurofisiologia. |
|||
:* [[Felis sylvestris catus]]<ref name="Lee-1995">{{Cite journal | last1 = Lee | first1 = A. | title = Animal models and vaccine development. | journal = Baillieres Clin Gastroenterol | volume = 9 | issue = 3 | pages = 615-32 | month = Sep | year = 1995 | doi = | PMID = 8563056 }}</ref><ref name="Meredith-1986">{{Cite journal | last1 = Meredith | first1 = MA. | last2 = Stein | first2 = BE. | title = Visual, auditory, and somatosensory convergence on cells in superior colliculus results in multisensory integration. | journal = J Neurophysiol | volume = 56 | issue = 3 | pages = 640-62 | month = Sep | year = 1986 | doi = | PMID = 3537225 }}</ref><ref name="Møller-1982">{{Cite journal | last1 = Møller | first1 = BR. | last2 = Mårdh | first2 = PA. | title = Animal models for the study of Chlamydial infections of the urogenital tract. | journal = Scand J Infect Dis Suppl | volume = 32 | issue = | pages = 103-8 | month = | year = 1982 | doi = | PMID = 6813962 }}</ref> - Il gatto è usato nelle ricerche di neurofisiologia. |
|||
==Organismi modello usati per specifiche ricerche == |
==Organismi modello usati per specifiche ricerche == |
||
Versione delle 10:57, 21 nov 2010

Un organismo modello è una specie estensivamente studiata per comprendere particolari fenomeni biologici, in base al presupposto che le acquisizioni fatte sull'organismo modello possano fornire indicazioni sugli altri organismi. Ciò è possibile grazie al fatto che i principi biologici fondamentali, come le vie metaboliche, di regolazione e di sviluppo, e i geni che le codificano, si mantengono attraverso l'evoluzione.
Il primo organismo modello impiegato in esperimenti rigorosi per la comprensione dell'ereditarietà è stato il Pisum sativum[1] di Gregor Mendel.[2][3] Il pisello da orto infatti risponde a specifiche esigenze di incrocio controllato, rapido passo generazionale, prole numerosa, caratteri fenotipici alternativi e disponibilità di numerose varietà commerciali. Queste caratteristiche lo resero ottimale per un approccio ai problemi della ereditarietà di tipo quantitativo e statistico.
Spesso, gli organismi modello vengono scelti in base alla loro capacità di essere adattabili a manipolazioni sperimentali. Di solito vengono preferite le seguenti caratteristiche: breve ciclo cellulare, tecniche per manipolazione genetica (ceppi inbred, linee di cellule staminali, e sistemi di transfezione). A volte, il riarrangiamento genetico favorisce il sequenziamento del genoma dell'organismo modello, per esempio, perché è molto compatto o per avere scarsa quantità di DNA non codificante, il cosiddetto "DNA spazzatura" (junk DNA).
Esistono numerosi organismi modello. Il primo organismo modello per la biologia molecolare probabilmente è stato il batterio E.coli, comunemente presente nel sistema digerente umano (e di solito ha attività benefica -- il pericoloso ceppo Escherichia coli O157:H7 è raro). Viene utilizzato anche nello studio di molti batteriofagi, specialmente il fago lambda.
Negli eucarioti sono stati studiati approfonditamente alcuni lieviti, specialmente il Saccharomyces cerevisiae (lievito della birra), soprattutto perché sono facili da gestire. Il ciclo cellulare in un lievito è molto simile al ciclo cellulare negli umani ed è regolato da proteine omologhe. È stato studiato anche il moscerino della frutta Drosophila melanogaster, sempre perché è facile da gestire per essere un organismo multicellulare. Il verme Caenorhabditis elegans è stato studiato perché ha stadi di sviluppo estremamente definiti ed è possibile, quindi, rivelare rapidamente delle anormalità.
Quando i ricercatori cercano un organismo da usare nei loro studi, prendono in considerazione parecchie caratteristiche. Le più comuni sono le dimensioni, il tempo di generazione, l'accessibilità, la manipolazione, la genetica, la conservazione dei meccanismi e un potenziale beneficio economico. Con la diffusione della biologia molecolare comparata, i ricercatori hanno cercato organismi modello che rappresentassero diverse tipologie di vita.
Principali organismi modello
Virus
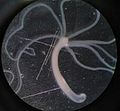
- Fago lambda[4][5] - batteriofago che infetta Escherichia coli
- Fago T4[6]
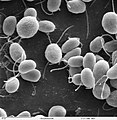
- Fago Phi-X174[7] E' stato il primo organismo seguenziato geneticamente.
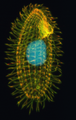
- virus del mosaico del tabacco[8][9][10]
-
Lisi di placche di E. coli da parte del batteriofago Lamda.
-
Struttura del capside del batteriofago phi X 174
-
Microscopia elettronica del Virus del Mosaico del Tabacco TMV
-
Virus del Mosaico del Tabacco a 160.000 X ingrandimenti
Procarioti
- Escherichia coli[11][12][13] - uno degli organismi unicellulari più studiati e più frequenti nell'intestino degli eucarioti superiori (ed in particolare di uccelli e mammiferi.
- Bacillus subtilis[14][15][16] - un batterio del suolo, ampiamente studiato per la sua produzione di spore.
- Mycoplasma genitalium[17][18][19] - un batterio parassita che vive nel tratto genitale e respiratorio dell'organismo umano.
- Vibrio fischeri[20][21] - organismo che vive in simbiosi con il calamaro gigante delle Hawaii (Euprymna scolopes)[20] poiché in grado di produrre la bioluminescenza necessaria al mollusco per illuminare l'ambiente circostante.
- Synechocystis[22][23][24] - un cianobatterio ampiamente usato nello studio della fotosintesi.
- Pseudomonas fluorescens[25][26][27] - un batterio del terreno in grado di diversificarsi velocemente in differenti linee cellulari.
Eucarioti unicellulari
- Saccharomyces cerevisiae[28][29][30] - noto come lievito della birra, è stato il primo eukaryota il cui genoma sia stato interamente sequenziato (1996).[31] Molto studiato per il suo ciclo cellulare.
- Schizosaccharomyces pombe[28][32][33] - altro modello di lievito, diverso da S.cerevisiae in particolare nella modalità di riproduzione: S.cerevisiae replica per gemmazione (budding), S.pombe per scissione binaria. Anche S.pombe è ampiamente studiato per il suo ciclo cellulare.
- Chlamydomonas reinhardtii[34][35][36] - un'alga verde unicellulare, utilizzata per studiare fotosintesi, flagelli e motilità, regolazione del metabolismo, riconoscimento cellula-cellula, risposta all'affamamento e diversi altri processi biologici. Ha una genetica molto studiata, con una grande quantità di mutanti noti e sequenziati e procedure relativamente semplici per produrne di nuovi. La sua crescita in laboratorio è infatti semplice e poco costosa. Esiste un centro di raccolta delle informazioni genetiche su Chamydomonas presso la Duke University e un gruppo internazionale di ricerca che tiene convegni periodici per discuterne.
- Tetrahymena thermophila[37][38][39] - un protozoo ciliato di acqua dolce che presenta dimorfismo del nucleo.
- Dictyostelium discoideum[40][41][42] - specie appartenente al genere Dictyostelium, molto studiata come modello in biologia dello sviluppo, biologia molecolare nonché in tossicologia ambientale.
Eucarioti pluricellulari
Piante
- Arabidopsis thaliana[28][43][44][45] - è probabilmente la più importante pianta modello. Il suo studio ha fornito una considerevole quantità di nuove conoscenze. È una specie a crescita veloce. È stata la prima pianta il cui genoma sia stato interamente sequenziato. (Biologia molecolare, Citologia)
- Riso (Oryza sativa)[46][47][48] - usato come modello per gli studi di biologia dei cereali. Il sequenziamento del suo genoma è in corso. (Agronomia, Biologia molecolare)
- Mais (Zea mais)[49][50][51] - modello per i cereali. Le sue caratteristiche genetiche hanno permesso lo sviluppo di teorie per la comprensione del ruolo dei trasposoni.[52] Il suo genoma è stato sequenziato. (Agronomia, Biologia molecolare)
- Tobacco BY-2 cells[53][54] - linea cellulare coltivata in sospensione della pianta di tabacco (Nicotiana tabacum). Viene utilizzata per studi generali per studi fisiologici a livello di biologia cellulare delle piante. Il genoma di questa particolare cultivar non sarà sequenziato (non almeno nell'immediato futuro), mentre quello della specie selvatica, Nicotiana tabaccum è attualmente in corso. (Biologia molecolare, Citologia)
- Lotus japonicus[55][56] - usato come modello per le piante leguminose (Agronomia, Biologia molecolare)
- Medicago truncatula[43][57][58] - altro modello per le leguminose. Il suo genoma, relativamente piccolo, è in sequenziamento. (Agronomia, Biologia molecolare)
- Brachypodium distachyon[59][60] - modello sperimentale emergente. Ha diversi attributi che lo rendono un eccellente modello per i cereali. (Agronomia, Biologia molecolare)
- Lemna gibba[61] É una pianta acquatica a rapida crescita, una delle più piccole piante da fiore. I tassi di crescita della Lemna sono utilizzati per valutare la tossicità dei prodotti chimici in ecotossicologia.
Funghi
Invertebrati
- Drosophila melanogaster[68][69][70] - il moscerino della frutta, uno dei più studiati organismi viventi. È stato utilizzato, sin dai primi decenni del 1900, per studi di genetica di base. Oggi è un ottimo modello animale per studi sul sistema nervoso[71][72][73] e sul suo sviluppatissimo sistema visivo.[74][75][76]
- Caenorhabditis elegans[77][78][79] - verme nematode, impiegato nello studio della differenziazione cellulare[80][81][82] e dell'apoptosi.[83][84][85]
- Hydra[86][87][88] - genere di organismi predatori di acqua dolce del phylum Cnidaria, dotati di simmetria radiale, molto studiati per l'alta capacità di rigenerazione.[89][90][91]
- Arbacia punctulata[92][93] - un riccio di mare della classe Echinoidea. Viene ancora ampiamente utilizzato come modello di embriogenesi.[94][95][96]
- Paracentrotus lividus[97][98][99] - un altro riccio di mare, anch'esso studiato da un punto di vista embriologico, ma anche da un punto di vista biomolecolare.
- Euprymna scolopes[100] - il calamaro gigante delle Hawaii, modello di simbiosi tra animali e batteri (in particolare il Vibrio fischeri[101][102]) nella produzione di sistemi bioluminescenti.[103]
- Loligo pealei[104][105][106] - un calamaro, oggetto di ampi studi neurologici a causa del suo "assone gigante" (quasi 1 mm di diamentro, circa 1000 volte maggiore di un assone medio di mammifero).
- Tribolium castaneum[107][108][109] - il tribolio delle farine, un piccolo insetto che cresce agevolmente, studiato soprattutto in studi sul comportamento.
- Ciona intestinalis[110][111][112] - l'ascidia lunga, ottimo organismo modello per studi sul genoma: ha un numero limitato di geni (circa 15000), un genoma compatto (circa 160 milioni di basi azotate) ed ha interessanti rapporti filogenetici con i vertebrati.
- Pristionchus pacificus[113][114][115] - verme cilindrico studiato nella biologia evolutiva dello sviluppo.
- Cepaea nemoralis[116][117][118] - mollusco gasteropode utilizzato negli studi sulla ereditarietà e in biologia evolutiva.
- Stomatogastric ganglion[119][120][121] il cui sistema digestivo è modello per la genesi di pattern motori.[122]
Vertebrati

- Canis lupus familiaris[123][124][125] - importante modello dei sistemi respiratorio e cardiovascolare.[126]
- Cavia porcellus[127][128][129] - la cavia, usata inizialmente da Robert Koch[130][131] e altri batteriologi, è diventata un sinonimo di "animale di laboratorio", per quanto oggi non sia più molto usata dalla ricerca.[129]
- Coturnix coturnix[132][133] - La quaglia viene utilizzata in esperimenti di embriologia perché presenta cellule colorate che permettono di utilizzarle in esperimenti di trapianto sul pollo.
- Cricetinae[134] - Il Criceto è usato nelle ricerche sulla leismaniosi[135]
- Gallus gallus[136][137][138] - Il pollo è particolarmente impiegato negli studi sullo sviluppo embrionale, in quanto facilmente maneggiabile e a rapido sviluppo.[139]
- Mus musculus[140] - il topo, modello animale più utilizzato nella ricerca biomedica. Ne esistono numerose linee inbred:[141][142][143] alcune sono state selezionate per mostrare particolari tratti, spesso di interesse medico, come il peso corporeo, la muscolatura, l'obesità.[144][145]
- Oryzias latipes[146][141][147] - un altro pesce, noto come medaka. Utilizzato anch'esso come modello di sviluppo, soprattutto dell'occhio, ha il vantaggio di essere più resistente di Zebrafish.[148]
- Rattus norvegicus[149][150][151] - il ratto, ampiamente usato come modello animale in tossicologia[152]; molto utile come modello neurologico e come fonte di colture primarie.[153]
- Sigmodon hispidus[154] [155] Usato nelle ricerche sulla poliomielite.
- Sus scrofa[156][157][158] - il maiale, modello animale utilizzato per studi pre-clinici di patologie come le retinopatie.[159][160][161] Una probabile applicazione futura è quella di modello per gene therapy.[162][163][164]
- Takifugu rubipres[165] - noto come pesce palla, ha un genoma molto compatto, con poco junk DNA.[166][167][168]
- Xenopus laevis[169][170][171] - un rospo africano, anch'esso molto usato in studi di biologia evolutiva dello sviluppo, soprattutto a causa delle dimensioni ampie delle sue cellule uovo.[172][173]
- Brachydanio rerio[174][175] o (Danio rerio) conosciuto come Zebrafish, è un pesce d'acqua dolce molto usato negli acquari. Ha un corpo quasi trasparente durante il primo sviluppo, ciò fornisce un accesso visivo all'anatomia interna dell'animale. Gli Zebrafish sono utilizzati per studiare lo sviluppo, la tossicologia[176] e tossicopatologia[177] e la specifica funzione del gene e il ruolo delle vie di segnalazione.
- Taeniopygia guttata[178][137][179] (Zebra finch) o (diamante mandarino viene usato per studi sugli apparati uditivi dei non mammiferi.
- Macaca mulatta[180] - Il Rhesus macaque o macaco vien usato negli studi sulla cognizione[181][182][183][184] e nelle malattie infettive.[185][186][187]
- Felis sylvestris catus[188][189][190] - Il gatto è usato nelle ricerche di neurofisiologia.
Organismi modello usati per specifiche ricerche
Selezione e conflitti sessuali
- Callosobruchus maculatus
- Chorthippus parallelus
- Coelopidae
- Diopsidae
- Drosophila spp.
- Macrostomum lignano
- Gryllus bimaculatus
- Scathophaga stercoraria
Zone ibride
Ecologia genomica
- Daphnia pulex, un organismo modello di indicatore comportamentale
Tavola genetica degli organismi modello
La tabella indica lo status del Progetto genoma per ciascun organismo, mostrando dell'organismo la ricombinazione omologa e lo stato delle conoscenze delle vie biochimiche dell'organismo.
| Organismo | Sequenziazione genomica | Ricombinazione omologa | Biochimica |
|---|---|---|---|
| Procariota | |||
| Escherichia coli | Si | Si | Eccellente |
| Eucariota unicellulare | |||
| Dictyostelium discoideum | Si | Si | Eccellente |
| Saccharomyces cerevisiae | Si | Si | Buono |
| Schizosaccharomyces pombe | Si | Si | Buono |
| Chlamydomonas reinhardtii | si | No | Buono |
| Tetrahymena thermophila | Si | Si | Buono |
| Eucariota unicellulare | |||
| Caenorhabditis elegans | Si | Sifficoltoso | Non così buono |
| Drosophila melanogaster | Si | Diffixile | Buono |
| Arabidopsis thaliana | Si | No | Cattivo |
| Physcomitrella patens | Si | Si | Eccellente |
| Vertebrato | |||
| Danio rerio | Si | No | Buono |
| Mus musculus | Si | Si | Buono |
| Xenopus laevis[191] | Si | ||
| Homo sapiens NB: non è un organismo modello | Si | Si | Buono |
Note
- ^ Update on the genetic control of flowering in garden pea., in J Exp Bot, vol. 60, n. 9, 2009, pp. 2493-9, DOI:10.1093/jxb/erp120.
- ^ The importance of starch biosynthesis in the wrinkled seed shape character of peas studied by Mendel., in Plant Mol Biol, vol. 22, n. 3, Jun 1993, pp. 525-31.
- ^ Fisher's contributions to genetics and heredity, with special emphasis on the Gregor Mendel controversy., in Biometrics, vol. 46, n. 4, Dec 1990, pp. 915-24.
- ^ [Bacteriophage lambda DNA replication--new discoveries made using an old experimental model], in Postepy Biochem, vol. 52, n. 1, 2006, pp. 42-8.
- ^ Methusaleh's Zoo: how nature provides us with clues for extending human health span., in J Comp Pathol, 142 Suppl 1, Jan 2010, pp. S10-21, DOI:10.1016/j.jcpa.2009.10.024.
- ^ Patterns of damage in genomic DNA sequences from a Neandertal., in Proc Natl Acad Sci U S A, vol. 104, n. 37, Sep 2007, pp. 14616-21, DOI:10.1073/pnas.0704665104.
- ^ Quantifying organismal complexity using a population genetic approach., in PLoS One, vol. 2, n. 2, 2007, pp. e217, DOI:10.1371/journal.pone.0000217.
- ^ Quantitative study of Au(III) and Pd(II) ion biosorption on genetically engineered Tobacco mosaic virus., in J Colloid Interface Sci, vol. 342, n. 2, Feb 2010, pp. 455-61, DOI:10.1016/j.jcis.2009.10.028.
- ^ From virus research to molecular biology: Tobacco mosaic virus in Germany, 1936-1956., in J Hist Biol, vol. 37, n. 2, 2004, pp. 259-301.
- ^ Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells., in Nucleic Acids Res, vol. 23, n. 9, May 1995, pp. 1557-60.
- ^ Mechanisms of recombination: lessons from E. coli., in Crit Rev Biochem Mol Biol, vol. 43, n. 6, pp. 347-70, DOI:10.1080/10409230802485358.
- ^ Integrative inference of gene-regulatory networks in Escherichia coli using information theoretic concepts and sequence analysis., in BMC Syst Biol, vol. 4, 2010, p. 116, DOI:10.1186/1752-0509-4-116.
- ^ Examination of genome homogeneity in prokaryotes using genomic signatures., in PLoS One, vol. 4, n. 12, 2009, pp. e8113, DOI:10.1371/journal.pone.0008113.
- ^ The interaction of Bacillus subtilis sigmaA with RNA polymerase., in Protein Sci, vol. 18, n. 11, Nov 2009, pp. 2287-97, DOI:10.1002/pro.239.
- ^ Identification of network topological units coordinating the global expression response to glucose in Bacillus subtilis and its comparison to Escherichia coli., in BMC Microbiol, vol. 9, 2009, p. 176, DOI:10.1186/1471-2180-9-176.
- ^ High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies., in PLoS Genet, vol. 4, n. 8, 2008, pp. e1000139, DOI:10.1371/journal.pgen.1000139.
- ^ MolliGen, a database dedicated to the comparative genomics of Mollicutes., in Nucleic Acids Res, vol. 32, Database issue, Jan 2004, pp. D307-10, DOI:10.1093/nar/gkh114.
- ^ Prolyl isomerases in a minimal cell. Catalysis of protein folding by trigger factor from Mycoplasma genitalium., in Eur J Biochem, vol. 267, n. 11, Jun 2000, pp. 3270-80.
- ^ A minimal gene set for cellular life derived by comparison of complete bacterial genomes., in Proc Natl Acad Sci U S A, vol. 93, n. 19, Sep 1996, pp. 10268-73.
- ^ a b Genetic analysis of trimethylamine N-oxide reductases in the light organ symbiont Vibrio fischeri ES114., in J Bacteriol, vol. 190, n. 17, Sep 2008, pp. 5814-23, DOI:10.1128/JB.00227-08.
- ^ Bacterial bioluminescence: organization, regulation, and application of the lux genes., in FASEB J, vol. 7, n. 11, Aug 1993, pp. 1016-22.
- ^ CyanoBase: the cyanobacteria genome database update 2010., in Nucleic Acids Res, vol. 38, Database issue, Jan 2010, pp. D379-81, DOI:10.1093/nar/gkp915.
- ^ Comparative genomics of NAD biosynthesis in cyanobacteria., in J Bacteriol, vol. 188, n. 8, Apr 2006, pp. 3012-23, DOI:10.1128/JB.188.8.3012-3023.2006.
- ^ A novel potassium channel in photosynthetic cyanobacteria., in PLoS One, vol. 5, n. 4, 2010, pp. e10118, DOI:10.1371/journal.pone.0010118.
- ^ A modeling and simulation study of siderophore mediated antagonism in dual-species biofilms., in Theor Biol Med Model, vol. 6, 2009, p. 30, DOI:10.1186/1742-4682-6-30.
- ^ Using Pseudomonas spp. for Integrated Biological Control., in Phytopathology, vol. 97, n. 2, Feb 2007, pp. 244-9, DOI:10.1094/PHYTO-97-2-0244.
- ^ Nitrogen availability to Pseudomonas fluorescens DF57 is limited during decomposition of barley straw in bulk soil and in the barley rhizosphere., in Appl Environ Microbiol, vol. 65, n. 10, Oct 1999, pp. 4320-8.
- ^ a b c The BioGRID Interaction Database: 2011 update., in Nucleic Acids Res, Nov 2010, DOI:10.1093/nar/gkq1116.
- ^ Discovery of Mutations in Saccharomyces cerevisiae by Pooled Linkage Analysis and Whole Genome Sequencing., in Genetics, Oct 2010, DOI:10.1534/genetics.110.123232.
- ^ Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes., in PLoS Comput Biol, vol. 6, n. 7, 2010, pp. e1000859, DOI:10.1371/journal.pcbi.1000859.
- ^ Life with 6000 genes., in Science, vol. 274, n. 5287, Oct 1996, pp. 546, 563-7.
- ^ Unconventional effects of UVA radiation on cell cycle progression in S. pombe., in Cell Cycle, vol. 7, n. 5, Mar 2008, pp. 611-22.
- ^ Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA., in Nat Struct Mol Biol, vol. 15, n. 1, Jan 2008, pp. 34-42, DOI:10.1038/nsmb1354.
- ^ Synthesizing and salvaging NAD: lessons learned from Chlamydomonas reinhardtii., in PLoS Genet, vol. 6, n. 9, 2010, DOI:10.1371/journal.pgen.1001105.
- ^ Origin of the polycomb repressive complex 2 and gene silencing by an E(z) homolog in the unicellular alga Chlamydomonas., in Epigenetics, vol. 5, n. 4, May 2010, pp. 301-12.
- ^ Flagellar elongation and gene expression in Chlamydomonas reinhardtii., in Eukaryot Cell, vol. 6, n. 8, Aug 2007, pp. 1411-20, DOI:10.1128/EC.00167-07.
- ^ Mutation accumulation in Tetrahymena., in BMC Evol Biol, vol. 10, n. 1, Nov 2010, p. 354, DOI:10.1186/1471-2148-10-354.
- ^ The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly., in Mol Biol Cell, vol. 20, n. 6, Mar 2009, pp. 1865-77, DOI:10.1091/mbc.E08-08-0838.
- ^ Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote., in PLoS Biol, vol. 4, n. 9, Sep 2006, pp. e286, DOI:10.1371/journal.pbio.0040286.
- ^ Centromere sequence and dynamics in Dictyostelium discoideum., in Nucleic Acids Res, vol. 37, n. 6, Apr 2009, pp. 1809-16, DOI:10.1093/nar/gkp017.
- ^ The actinome of Dictyostelium discoideum in comparison to actins and actin-related proteins from other organisms., in PLoS One, vol. 3, n. 7, 2008, pp. e2654, DOI:10.1371/journal.pone.0002654.
- ^ Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium., in BMC Genomics, vol. 9, 2008, p. 291, DOI:10.1186/1471-2164-9-291.
- ^ a b Genome-wide investigation reveals high evolutionary rates in annual model plants., in BMC Plant Biol, vol. 10, n. 1, Nov 2010, p. 242, DOI:10.1186/1471-2229-10-242.
- ^ Identification and structural characterization of FYVE domain-containing proteins of Arabidopsis thaliana., in BMC Plant Biol, vol. 10, 2010, p. 157, DOI:10.1186/1471-2229-10-157.
- ^ The evolution of a high copy gene array in Arabidopsis., in J Mol Evol, vol. 70, n. 6, Jun 2010, pp. 531-44, DOI:10.1007/s00239-010-9350-2.
- ^ Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses., in Proc Natl Acad Sci U S A, vol. 102, n. 37, Sep 2005, pp. 13206-11, DOI:10.1073/pnas.0502365102.
- ^ Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses., in Plant Physiol, vol. 135, n. 3, Jul 2004, pp. 1776-83, DOI:10.1104/pp.104.039818.
- ^ Characterization of paralogous protein families in rice., in BMC Plant Biol, vol. 8, 2008, p. 18, DOI:10.1186/1471-2229-8-18.
- ^ Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis., in Plant Cell, vol. 14, n. 12, Dec 2002, pp. 3119-32.
- ^ The characterization of ligand-specific maize (Zea mays) profilin mutants., in Biochem J, vol. 358, Pt 1, Aug 2001, pp. 49-57.
- ^ Intron-exon structures of eukaryotic model organisms., in Nucleic Acids Res, vol. 27, n. 15, Aug 1999, pp. 3219-28.
- ^ Transposon excision from an atypical site: a mechanism of evolution of novel transposable elements., in PLoS One, vol. 2, n. 10, 2007, pp. e965, DOI:10.1371/journal.pone.0000965.
- ^ Plant cell division is specifically affected by nitrotyrosine., in J Exp Bot, vol. 61, n. 3, Mar 2010, pp. 901-9, DOI:10.1093/jxb/erp369.
- ^ Passage of Trojan peptoids into plant cells., in Chembiochem, vol. 10, n. 15, Oct 2009, pp. 2504-12, DOI:10.1002/cbic.200900331.
- ^ Lotus genome: pod of gold for legume research., in Trends Plant Sci, vol. 13, n. 10, Oct 2008, pp. 515-7, DOI:10.1016/j.tplants.2008.08.001.
- ^ Functional characterization of an unusual phytochelatin synthase, LjPCS3, of Lotus japonicus., in Plant Physiol, vol. 148, n. 1, Sep 2008, pp. 536-45, DOI:10.1104/pp.108.121715.
- ^ Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds., in Plant Physiol, vol. 144, n. 3, Jul 2007, pp. 1559-79, DOI:10.1104/pp.107.098103.
- ^ MtDB: a database for personalized data mining of the model legume Medicago truncatula transcriptome., in Nucleic Acids Res, vol. 31, n. 1, Jan 2003, pp. 196-201.
- ^ Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon., in BMC Genomics, vol. 11, 2010, p. 600, DOI:10.1186/1471-2164-11-600.
- ^ Conserved microRNAs and their targets in model grass species Brachypodium distachyon., in Planta, vol. 230, n. 4, Sep 2009, pp. 659-69, DOI:10.1007/s00425-009-0974-7.
- ^ Monensin is not toxic to aquatic macrophytes at environmentally relevant concentrations., in Arch Environ Contam Toxicol, vol. 53, n. 4, Nov 2007, pp. 541-51, DOI:10.1007/s00244-007-0002-5.
- ^ Study of codon bias perspective of fungal xylanase gene by multivariate analysis., in Bioinformation, vol. 3, n. 10, 2009, pp. 425-9.
- ^ The glutathione system of Aspergillus nidulans involves a fungus-specific glutathione S-transferase., in J Biol Chem, vol. 284, n. 12, Mar 2009, pp. 8042-53, DOI:10.1074/jbc.M807771200.
- ^ Systems analysis unfolds the relationship between the phosphoketolase pathway and growth in Aspergillus nidulans., in PLoS One, vol. 3, n. 12, 2008, pp. e3847, DOI:10.1371/journal.pone.0003847.
- ^ Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa., in Proc Natl Acad Sci U S A, vol. 106, n. 52, Dec 2009, pp. 22157-62, DOI:10.1073/pnas.0906810106.
- ^ SnoRNAs from the filamentous fungus Neurospora crassa: structural, functional and evolutionary insights., in BMC Genomics, vol. 10, 2009, p. 515, DOI:10.1186/1471-2164-10-515.
- ^ Neurospora as a model fungus for studies in cytogenetics and sexual biology at Stanford., in J Biosci, vol. 34, n. 1, Mar 2009, pp. 139-59.
- ^ Drosophila Genetic Resource and Stock Center; The National BioResource Project., in Exp Anim, vol. 59, n. 2, 2010, pp. 125-38.
- ^ Lessons from a compartmental model of a Drosophila neuron., in J Neurosci, vol. 29, n. 39, Sep 2009, pp. 12033-4, DOI:10.1523/JNEUROSCI.3348-09.2009.
- ^ Recent advances in Drosophila stem cell biology., in Int J Dev Biol, vol. 53, n. 8-10, 2009, pp. 1329-39, DOI:10.1387/ijdb.072431jp.
- ^ Molecular and cellular mechanisms of lamina-specific axon targeting., in Cold Spring Harb Perspect Biol, vol. 2, n. 3, Mar 2010, pp. a001743, DOI:10.1101/cshperspect.a001743.
- ^ Insulin/IGF-like signalling, the central nervous system and aging., in Biochem J, vol. 418, n. 1, Feb 2009, pp. 1-12, DOI:10.1042/BJ20082102.
- ^ Dissection of larval CNS in Drosophila melanogaster., in J Vis Exp, n. 1, Dec 2006, p. 85, DOI:10.3791/85.
- ^ The gang of four gene regulates growth and patterning of the developing Drosophila eye., in Fly (Austin), vol. 4, n. 2, Apr 2010, pp. 104-16.
- ^ Dystroglycan and mitochondrial ribosomal protein l34 regulate differentiation in the Drosophila eye., in PLoS One, vol. 5, n. 5, 2010, pp. e10488, DOI:10.1371/journal.pone.0010488.
- ^ Cell cycle arrest by a gradient of Dpp signaling during Drosophila eye development., in BMC Dev Biol, vol. 10, 2010, p. 28, DOI:10.1186/1471-213X-10-28.
- ^ C. elegans Mutant Identification with a One-Step Whole-Genome-Sequencing and SNP Mapping Strategy., in PLoS One, vol. 5, n. 11, 2010, pp. e15435, DOI:10.1371/journal.pone.0015435.
- ^ Neurogenesis in the nematode Caenorhabditis elegans., in WormBook, 2010, pp. 1-24, DOI:10.1895/wormbook.1.12.2.
- ^ Trans-generational epigenetic regulation of C. elegans primordial germ cells., in Epigenetics Chromatin, vol. 3, n. 1, 2010, p. 15, DOI:10.1186/1756-8935-3-15.
- ^ microRNA expression patterns reveal differential expression of target genes with age., in PLoS One, vol. 5, n. 5, 2010, pp. e10724, DOI:10.1371/journal.pone.0010724.
- ^ Genome-wide gene expression regulation as a function of genotype and age in C. elegans., in Genome Res, vol. 20, n. 7, Jul 2010, pp. 929-37, DOI:10.1101/gr.102160.109.
- ^ Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network., in Mol Syst Biol, vol. 6, May 2010, p. 367, DOI:10.1038/msb.2010.23.
- ^ Structural and biochemical characterization of CRN-5 and Rrp46: an exosome component participating in apoptotic DNA degradation., in RNA, vol. 16, n. 9, Sep 2010, pp. 1748-59, DOI:10.1261/rna.2180810.
- ^ The apoptosome at high resolution., in Cell, vol. 141, n. 3, Apr 2010, pp. 402-4, DOI:10.1016/j.cell.2010.04.015.
- ^ Radiation biology of Caenorhabditis elegans: germ cell response, aging and behavior., in J Radiat Res (Tokyo), vol. 51, n. 2, Mar 2010, pp. 107-21.
- ^ Axial patterning in hydra., in Cold Spring Harb Perspect Biol, vol. 1, n. 1, Jul 2009, pp. a000463, DOI:10.1101/cshperspect.a000463.
- ^ Evolutionary history of the HAP2/GCS1 gene and sexual reproduction in metazoans., in PLoS One, vol. 4, n. 11, 2009, pp. e7680, DOI:10.1371/journal.pone.0007680.
- ^ A novel gene family controls species-specific morphological traits in Hydra., in PLoS Biol, vol. 6, n. 11, Nov 2008, pp. e278, DOI:10.1371/journal.pbio.0060278.
- ^ Models for the generation and interpretation of gradients., in Cold Spring Harb Perspect Biol, vol. 1, n. 4, Oct 2009, pp. a001362, DOI:10.1101/cshperspect.a001362.
- ^ Hydra regeneration and epitheliopeptides., in Dev Dyn, vol. 226, n. 2, Feb 2003, pp. 182-9, DOI:10.1002/dvdy.10221.
- ^ HyAlx, an aristaless-related gene, is involved in tentacle formation in hydra., in Development, vol. 127, n. 22, Nov 2000, pp. 4743-52.
- ^ Modulation of the development of plutei by nitric oxide in the sea urchin Arbacia punctulata., in Biol Bull, vol. 199, n. 2, Oct 2000, pp. 195-7.
- ^ Ryanodine-sensitive calcium flux regulates motility of Arbacia punctulata sperm., in Biol Bull, vol. 205, n. 2, Oct 2003, pp. 185-6.
- ^ [Influence of NO-synthase inhibitors on embryonal development of sea urchins.], in Zh Evol Biokhim Fiziol, vol. 40, n. 3, pp. 229-34.
- ^ An ultracytochemical study of the respiratory potency, integrity, and fate of the sea urchin sperm mitochondria during early embryogenesis., in J Cell Biol, vol. 66, n. 2, Aug 1975, pp. 367-76.
- ^ Induction of a reductive pathway for deoxyribonucleotide synthesis during early embryogenesis of the sea urchin., in Proc Natl Acad Sci U S A, vol. 69, n. 8, Aug 1972, pp. 2006-10.
- ^ Assessment of DNA damage by RAPD in Paracentrotus lividus embryos exposed to amniotic fluid from residents living close to waste landfill sites., in J Biomed Biotechnol, vol. 2010, 2010, DOI:10.1155/2010/251767.
- ^ A method for measuring mitochondrial mass and activity., in Cytotechnology, vol. 56, n. 3, Mar 2008, pp. 145-9, DOI:10.1007/s10616-008-9143-2.
- ^ First cell cycles of sea urchin Paracentrotus lividus are dramatically impaired by exposure to extremely low-frequency electromagnetic field., in Biol Reprod, vol. 75, n. 6, Dec 2006, pp. 948-53, DOI:10.1095/biolreprod.106.051227.
- ^ Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from Two Oahu (Hawaii) populations., in Appl Environ Microbiol, vol. 75, n. 1, Jan 2009, pp. 193-202, DOI:10.1128/AEM.01792-08.
- ^ H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri., in Proc Natl Acad Sci U S A, vol. 107, n. 18, May 2010, pp. 8375-80, DOI:10.1073/pnas.1003571107.
- ^ Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis., in Proc Natl Acad Sci U S A, vol. 107, n. 5, Feb 2010, pp. 2259-64, DOI:10.1073/pnas.0909712107.
- ^ Evidence for light perception in a bioluminescent organ., in Proc Natl Acad Sci U S A, vol. 106, n. 24, Jun 2009, pp. 9836-41, DOI:10.1073/pnas.0904571106.
- ^ Squid (Loligo pealei) giant fiber system: a model for studying neurodegeneration and dementia?, in Biol Bull, vol. 210, n. 3, Jun 2006, pp. 318-33.
- ^ Vesicular reuptake inhibition by a synaptotagmin I C2B domain antibody at the squid giant synapse., in Proc Natl Acad Sci U S A, vol. 101, n. 51, Dec 2004, pp. 17855-60, DOI:10.1073/pnas.0408200101.
- ^ Crystallization and preliminary X-ray diffraction analysis of calexcitin from Loligo pealei: a neuronal protein implicated in learning and memory., in Acta Crystallogr Sect F Struct Biol Cryst Commun, vol. 61, Pt 10, Oct 2005, pp. 879-81, DOI:10.1107/S1744309105026758.
- ^ 3D Standard Brain of the Red Flour Beetle Tribolium Castaneum: A Tool to Study Metamorphic Development and Adult Plasticity., in Front Syst Neurosci, vol. 4, 2010, p. 3, DOI:10.3389/neuro.06.003.2010.
- ^ Forward genetics in Tribolium castaneum: opening new avenues of research in arthropod biology., in J Biol, vol. 8, n. 12, 2009, p. 106, DOI:10.1186/jbiol208.
- ^ BeetleBase in 2010: revisions to provide comprehensive genomic information for Tribolium castaneum., in Nucleic Acids Res, vol. 38, Database issue, Jan 2010, pp. D437-42, DOI:10.1093/nar/gkp807.
- ^ The transcription/migration interface in heart precursors of Ciona intestinalis., in Science, vol. 320, n. 5881, Jun 2008, pp. 1349-52, DOI:10.1126/science.1158170.
- ^ Similar regulatory logic in Ciona intestinalis for two Wnt pathway modulators, ROR and SFRP-1/5., in Dev Biol, vol. 329, n. 2, May 2009, pp. 364-73, DOI:10.1016/j.ydbio.2009.02.018.
- ^ Delineating metamorphic pathways in the ascidian Ciona intestinalis., in Dev Biol, vol. 326, n. 2, Feb 2009, pp. 357-67, DOI:10.1016/j.ydbio.2008.11.026.
- ^ Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models., in Genome Res, vol. 20, n. 6, Jun 2010, pp. 837-46, DOI:10.1101/gr.103119.109.
- ^ Repertoire and evolution of miRNA genes in four divergent nematode species., in Genome Res, vol. 19, n. 11, Nov 2009, pp. 2064-74, DOI:10.1101/gr.093781.109.
- ^ Natural variation of outcrossing in the hermaphroditic nematode Pristionchus pacificus., in BMC Evol Biol, vol. 9, 2009, p. 75, DOI:10.1186/1471-2148-9-75.
- ^ Disequilibrium in some Cepaea populations., in Heredity, vol. 94, n. 5, May 2005, pp. 497-500, DOI:10.1038/sj.hdy.6800645.
- ^ History or current selection? A molecular analysis of 'area effects' in the land snail Cepaea nemoralis., in Proc Biol Sci, vol. 267, n. 1451, Jul 2000, pp. 1399-405, DOI:10.1098/rspb.2000.1156.
- ^ Spatial structure of shell polychromatism in populations of Cepaea nemoralis: new techniques for an old debate., in Heredity, vol. 88, n. 1, Jan 2002, pp. 75-82, DOI:10.1038/sj.hdy.6800012.
- ^ Precise temperature compensation of phase in a rhythmic motor pattern., in PLoS Biol, vol. 8, n. 8, 2010, DOI:10.1371/journal.pbio.1000469.
- ^ Hormonal modulation of sensorimotor integration., in J Neurosci, vol. 30, n. 7, Feb 2010, pp. 2418-27, DOI:10.1523/JNEUROSCI.5533-09.2010.
- ^ Neural mechanisms underlying the generation of the lobster gastric mill motor pattern., in Front Neural Circuits, vol. 3, 2009, p. 12, DOI:10.3389/neuro.04.012.2009.
- ^ Functional consequences of animal-to-animal variation in circuit parameters., in Nat Neurosci, vol. 12, n. 11, Nov 2009, pp. 1424-30, DOI:10.1038/nn.2404.
- ^ (EN) www.official-documents.gov.uk (PDF), su official-documents.gov.uk.
- ^ (EN) Animal-Testing Ban for Cosmetics to Be Upheld, EU Court Rules - Bloomberg, su bloomberg.com.
- ^ (EN) US FDA/CFSAN - Animal Testing, su cfsan.fda.gov (archiviato dall'url originale il 23 maggio 2007).
- ^ Safar P, Behringer W, Böttiger BW, Sterz F, Cerebral resuscitation potentials for cardiac arrest, in Crit. Care Med., vol. 30, 4 Suppl, April 2002, pp. S140–4.
- ^ Ototoxic drugs: difference in sensitivity between mice and guinea pigs., in Toxicol Lett, vol. 193, n. 1, Mar 2010, pp. 41-9, DOI:10.1016/j.toxlet.2009.12.003.
- ^ Identification, sequencing, and cellular localization of hepcidin in guinea pig (Cavia porcellus)., in J Endocrinol, vol. 202, n. 3, Sep 2009, pp. 389-96, DOI:10.1677/JOE-09-0191.
- ^ a b Preference, adaptation and survival of Mycoplasma pneumoniae subtypes in an animal model., in Int J Med Microbiol, vol. 294, n. 2-3, Sep 2004, pp. 149-55.
- ^ Experimental models used to study human tuberculosis., in Adv Appl Microbiol, vol. 71, 2010, pp. 75-89, DOI:10.1016/S0065-2164(10)71003-0.
- ^ Past, present and future directions in human genetic susceptibility to tuberculosis., in FEMS Immunol Med Microbiol, vol. 58, n. 1, Feb 2010, pp. 3-26, DOI:10.1111/j.1574-695X.2009.00600.x.
- ^ Quail Genomics: a knowledgebase for Northern bobwhite., in BMC Bioinformatics, 11 Suppl 6, 2010, pp. S13, DOI:10.1186/1471-2105-11-S6-S13.
- ^ The effect of different foam concentrations on sperm motility in Japanese quail., in Vet Med Int, vol. 2010, 2010, p. 564921, DOI:10.4061/2010/564921.
- ^ The hamster handbook, Hauppauge, NY, Barron's, 2003, ISBN 978-0-7641-2294-1.
- ^ Hamster model of leptospirosis., in Curr Protoc Microbiol, Chapter 12, Sep 2006, pp. Unit 12E.2, DOI:10.1002/9780471729259.mc12e02s02.
- ^ Selection for the compactness of highly expressed genes in Gallus gallus., in Biol Direct, vol. 5, 2010, p. 35, DOI:10.1186/1745-6150-5-35.
- ^ a b The genome of a songbird., in Nature, vol. 464, n. 7289, Apr 2010, pp. 757-62, DOI:10.1038/nature08819.
- ^ Red jungle fowl (Gallus gallus) as a model for studying the molecular mechanism of seasonal reproduction., in Anim Sci J, vol. 80, n. 3, Jun 2009, pp. 328-32, DOI:10.1111/j.1740-0929.2009.00628.x.
- ^ [Interrelationship between NAD metabolism and DNA synthesis in chicken liver nuclei during ontogenesis], in Biokhimiia, vol. 42, n. 2, Feb 1977, pp. 251-6.
- ^ The transformation of the model organism: a decade of developmental genetics., in Nat Genet, 33 Suppl, Mar 2003, pp. 285-93, DOI:10.1038/ng1105.
- ^ a b The National BioResource Project Medaka (NBRP Medaka): an integrated bioresource for biological and biomedical sciences., in Exp Anim, vol. 59, n. 1, 2010, pp. 13-23.
- ^ Murine models of human acute myeloid leukemia., in Cancer Treat Res, vol. 145, 2010, pp. 183-96, DOI:10.1007/978-0-387-69259-3_11.
- ^ Mouse models with human immunity and their application in biomedical research., in J Cell Mol Med, vol. 13, n. 6, Jun 2009, pp. 1043-58, DOI:10.1111/j.1582-4934.2008.00347.x.
- ^ Loci on chromosomes 2, 4, 9, and 16 for body weight, body length, and adiposity identified in a genome scan of an F2 intercross between the 129P3/J and C57BL/6ByJ mouse strains., in Mamm Genome, vol. 14, n. 5, May 2003, pp. 302-13.
- ^ Dose-response effects of ectopic agouti protein on iron overload and age-associated aspects of the Avy/a obese mouse phenome., in Peptides, vol. 26, n. 10, Oct 2005, pp. 1697-711, DOI:10.1016/j.peptides.2004.12.033.
- ^ Quantifiable biomarkers of normal aging in the Japanese medaka fish (Oryzias latipes)., in PLoS One, vol. 5, n. 10, 2010, pp. e13287, DOI:10.1371/journal.pone.0013287.
- ^ The Japanese medaka, Oryzias latipes, as a new model organism for studying environmental germ-cell mutagenesis., in Environ Health Perspect, 102 Suppl 12, Dec 1994, pp. 33-5.
- ^ Cloning and developmental expression of kinesin superfamily7 (kif7) in the brackish medaka (Oryzias melastigma), a close relative of the Japanese medaka (Oryzias latipes)., in Mar Pollut Bull, vol. 57, n. 6-12, 2008, pp. 425-32, DOI:10.1016/j.marpolbul.2008.02.044.
- ^ Evaluation of neonatally-induced mild diabetes in rats: Maternal and fetal repercussions., in Diabetol Metab Syndr, vol. 2, n. 1, 2010, p. 37, DOI:10.1186/1758-5996-2-37.
- ^ The Rat Genome Database 2009: variation, ontologies and pathways., in Nucleic Acids Res, vol. 37, Database issue, Jan 2009, pp. D744-9, DOI:10.1093/nar/gkn842.
- ^ Rapid but not slow spinal cord compression elicits neurogenic pulmonary edema in the rat., in Physiol Res, vol. 58, n. 2, 2009, pp. 269-77.
- ^ Methylated arsenicals: the implications of metabolism and carcinogenicity studies in rodents to human risk assessment., in Crit Rev Toxicol, vol. 36, n. 2, Feb 2006, pp. 99-133.
- ^ Novel approaches to models of Alzheimer's disease pathology for drug screening and development., in J Mol Neurosci, vol. 24, n. 1, 2004, pp. 23-32, DOI:10.1385/JMN:24:1:023.
- ^ Mammal species of the world: a taxonomic and geographic reference, Baltimore, Johns Hopkins University Press, 2005, ISBN 978-0-8018-8221-0.
- ^ The marmoset and cotton rat as animal models for the study of sperm chromatin packaging., in Syst Biol Reprod Med, vol. 56, n. 3, Jun 2010, pp. 207-12, DOI:10.3109/19396361003653311.
- ^ Genetic resources, genome mapping and evolutionary genomics of the pig (Sus scrofa)., in Int J Biol Sci, vol. 3, n. 3, 2007, pp. 153-65.
- ^ Transcriptome architecture across tissues in the pig., in BMC Genomics, vol. 9, 2008, p. 173, DOI:10.1186/1471-2164-9-173.
- ^ Cloning, characterization and expression analysis of porcine microRNAs., in BMC Genomics, vol. 10, 2009, p. 65, DOI:10.1186/1471-2164-10-65.
- ^ Mitogen-activated protein kinases in the porcine retinal arteries and neuroretina following retinal ischemia-reperfusion., in Mol Vis, vol. 16, 2010, pp. 392-407.
- ^ Immunohistochemical study of pig retinal development., in Mol Vis, vol. 15, 2009, pp. 1915-28.
- ^ Effect of systemic nitric oxide synthase inhibition on optic disc oxygen partial pressure in normoxia and in hypercapnia., in Invest Ophthalmol Vis Sci, vol. 50, n. 1, Jan 2009, pp. 378-84, DOI:10.1167/iovs.08-2413.
- ^ Therapy of experimental type 1 diabetes by isolated Sertoli cell xenografts alone., in J Exp Med, vol. 206, n. 11, Oct 2009, pp. 2511-26, DOI:10.1084/jem.20090134.
- ^ Cell-based resurfacing of large cartilage defects: long-term evaluation of grafts from autologous transgene-activated periosteal cells in a porcine model of osteoarthritis., in Arthritis Rheum, vol. 58, n. 2, Feb 2008, pp. 475-88, DOI:10.1002/art.23124.
- ^ Morphological characteristics of submandibular glands of miniature pig., in Chin Med J (Engl), vol. 118, n. 16, Aug 2005, pp. 1368-73.
- ^ Explosive speciation of Takifugu: another use of fugu as a model system for evolutionary biology., in Mol Biol Evol, vol. 26, n. 3, Mar 2009, pp. 623-9, DOI:10.1093/molbev/msn283.
- ^ Investigation of loss and gain of introns in the compact genomes of pufferfishes (Fugu and Tetraodon)., in Mol Biol Evol, vol. 25, n. 3, Mar 2008, pp. 526-35, DOI:10.1093/molbev/msm278.
- ^ The genome size evolution of medaka (Oryzias latipes) and fugu (Takifugu rubripes)., in Genes Genet Syst, vol. 82, n. 2, Apr 2007, pp. 135-44.
- ^ DED: Database of Evolutionary Distances., in Nucleic Acids Res, vol. 33, Database issue, Jan 2005, pp. D442-6, DOI:10.1093/nar/gki094.
- ^ Xenopus pancreas development., in Dev Dyn, vol. 238, n. 6, Jun 2009, pp. 1271-86, DOI:10.1002/dvdy.21935.
- ^ An ontology for Xenopus anatomy and development., in BMC Dev Biol, vol. 8, 2008, p. 92, DOI:10.1186/1471-213X-8-92.
- ^ Manipulating the Xenopus genome with transposable elements., in Genome Biol, 8 Suppl 1, 2007, pp. S11, DOI:10.1186/gb-2007-8-s1-s11.
- ^ Xenbase: a Xenopus biology and genomics resource., in Nucleic Acids Res, vol. 36, Database issue, Jan 2008, pp. D761-7, DOI:10.1093/nar/gkm826.
- ^ Xenopus as a model organism in developmental chemical genetic screens., in Mol Biosyst, vol. 1, n. 3, Sep 2005, pp. 223-8, DOI:10.1039/b506103b.
- ^ Special feature--roundtable discussion. Fish models for studying adaptive evolution and speciation., in Zebrafish, vol. 2, n. 3, 2005, pp. 147-56, DOI:10.1089/zeb.2005.2.147.
- ^ Small fish, big science., in PLoS Biol, vol. 2, n. 5, May 2004, pp. E148, DOI:10.1371/journal.pbio.0020148.
- ^ Zebrafish as a model vertebrate for investigating chemical toxicity., in Toxicol Sci, vol. 86, n. 1, Jul 2005, pp. 6-19, DOI:10.1093/toxsci/kfi110.
- ^ The state of the art of the zebrafish model for toxicology and toxicologic pathology research--advantages and current limitations., in Toxicol Pathol, 31 Suppl, pp. 62-87.
- ^ Diversity and temporal stability of bacterial communities in a model passerine bird, the zebra finch., in Mol Ecol, Nov 2010, DOI:10.1111/j.1365-294X.2010.04892.x.
- ^ Utilization of a zebra finch BAC library to determine the structure of an avian androgen receptor genomic region., in Genomics, vol. 87, n. 1, Jan 2006, pp. 181-90, DOI:10.1016/j.ygeno.2005.09.005.
- ^ Animal models of human microsporidial infections., in Lab Anim Sci, vol. 48, n. 6, Dec 1998, pp. 589-92.
- ^ Global image dissimilarity in macaque inferotemporal cortex predicts human visual search efficiency., in J Neurosci, vol. 30, n. 4, Jan 2010, pp. 1258-69, DOI:10.1523/JNEUROSCI.1908-09.2010.
- ^ Cooperative and competitive interactions facilitate stereo computations in macaque primary visual cortex., in J Neurosci, vol. 29, n. 50, Dec 2009, pp. 15780-95, DOI:10.1523/JNEUROSCI.2305-09.2009.
- ^ Spatial decisions and cognitive strategies of monkeys and humans based on abstract spatial stimuli in rotation test., in Proc Natl Acad Sci U S A, vol. 106, n. 36, Sep 2009, pp. 15478-82, DOI:10.1073/pnas.0907053106.
- ^ Thatcher effect in monkeys demonstrates conservation of face perception across primates., in Curr Biol, vol. 19, n. 15, Aug 2009, pp. 1270-3, DOI:10.1016/j.cub.2009.05.067.
- ^ Is an HIV vaccine possible?, in Braz J Infect Dis, vol. 13, n. 4, Aug 2009, pp. 304-10.
- ^ Immunity in natural SIV infections., in J Intern Med, vol. 265, n. 1, Jan 2009, pp. 97-109, DOI:10.1111/j.1365-2796.2008.02049.x.
- ^ The utility of rhesus monkey (Macaca mulatta) and other non-human primate models for preclinical testing of Leishmania candidate vaccines., in Mem Inst Oswaldo Cruz, vol. 103, n. 7, Nov 2008, pp. 629-44.
- ^ Animal models and vaccine development., in Baillieres Clin Gastroenterol, vol. 9, n. 3, Sep 1995, pp. 615-32.
- ^ Visual, auditory, and somatosensory convergence on cells in superior colliculus results in multisensory integration., in J Neurophysiol, vol. 56, n. 3, Sep 1986, pp. 640-62.
- ^ Animal models for the study of Chlamydial infections of the urogenital tract., in Scand J Infect Dis Suppl, vol. 32, 1982, pp. 103-8.
- ^ JGI-Led Team Sequences Frog Genome, in GenomeWeb.com, Genome Web, 29 aprile 2010. URL consultato il 30 aprile 2010.
Bibliografia
- (EN) Spitsbergen J.M. and Kent M.L. (2003). The state of the art of the zebrafish model for toxicology and toxicologic pathology research--advantages and current limitations. Toxicol Pathol. 31 (Supplement), 62-87. PubMed Abstract Link => PMID 12597434.Testo su Zebrafish disponibile gratuitamente.
- (EN) Riddle, Donald L.; Blumenthal, Thomas; Meyer, Barbara J.; and Priess, James R. (Eds.). (1997). C. ELEGANS II. Woodbury, NY: Cold Spring Harbor Press. ISBN 0-87969-532-3. Testo su C.elegans disponibile gratuitamente.
- Mammal species of the world: a taxonomic and geographic reference, Baltimore, Johns Hopkins University Press, 2005, ISBN 978-0-8018-8221-0.
- The hamster handbook, Hauppauge, NY, Barron's, 2003, ISBN 978-0-7641-2294-1.
Voci correlate
Collegamenti esterni
- (EN) Banca dati contenente i genomi di tutti gli organismi sequenziati
- (EN) Banca dati NCBI contenente informazioni riguardanti tutti gli organismi modello
- (EN) Altra banca dati contenente informazioni su vari organismi modello
- (EN) Wellcome Trust description of model organisms
- (EN) Model organisms in developmental biology
- (EN) Ludwig-Maximillians-Universität Department Biologie II
- (EN) Zebrafish GenomeWiki Community Annotation Project mantenuto dall'Institute of Genomics and Integrative Biology
- (EN) Workhorse Zoo by Adam Zaretsky










![Zea mays]](http://upload.wikimedia.org/wikipedia/commons/thumb/3/32/Maispflanze.jpg/90px-Maispflanze.jpg)